Motivation for isotwas
motivation.Rmdisotwas provides functions to run transcriptome-wide
association studies on the isoform-level.
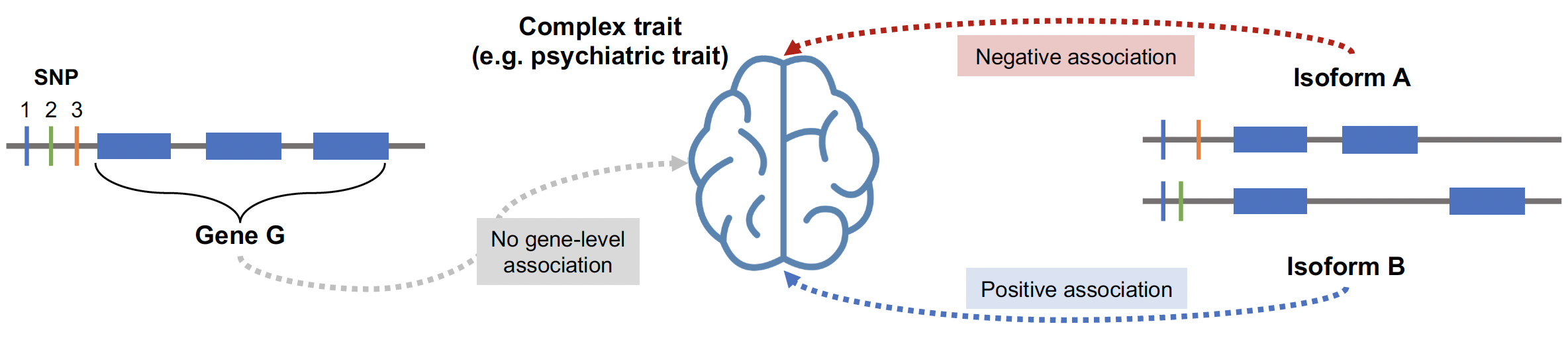
Isoform-level analyses can provide further granularity to a gene-trait association by pinpointing the isoform of a given gene that drives the association. Furthermore, if two isoforms of the same genes have associations with divergent effect sizes, then gene-level trait mapping will likely miss this association but isoform-level trait mapping will not.
isotwas contains functions for:
-
Training multivariate predictive models for isoform-level expression from genetic variants, with 9 different methods including:
- MRCE (Multivariate Regression with Covariance Estimation)
- Multivariate elastic net
- Joinet stacked elastic net
- Sparse PLS
- Sparse group lasso
- Multi-task lasso (L21 regularization)
- Super learner stacking
- Graph-regularized regression (using isoform similarity)
- Univariate methods (elastic net, BLUP, SuSiE)
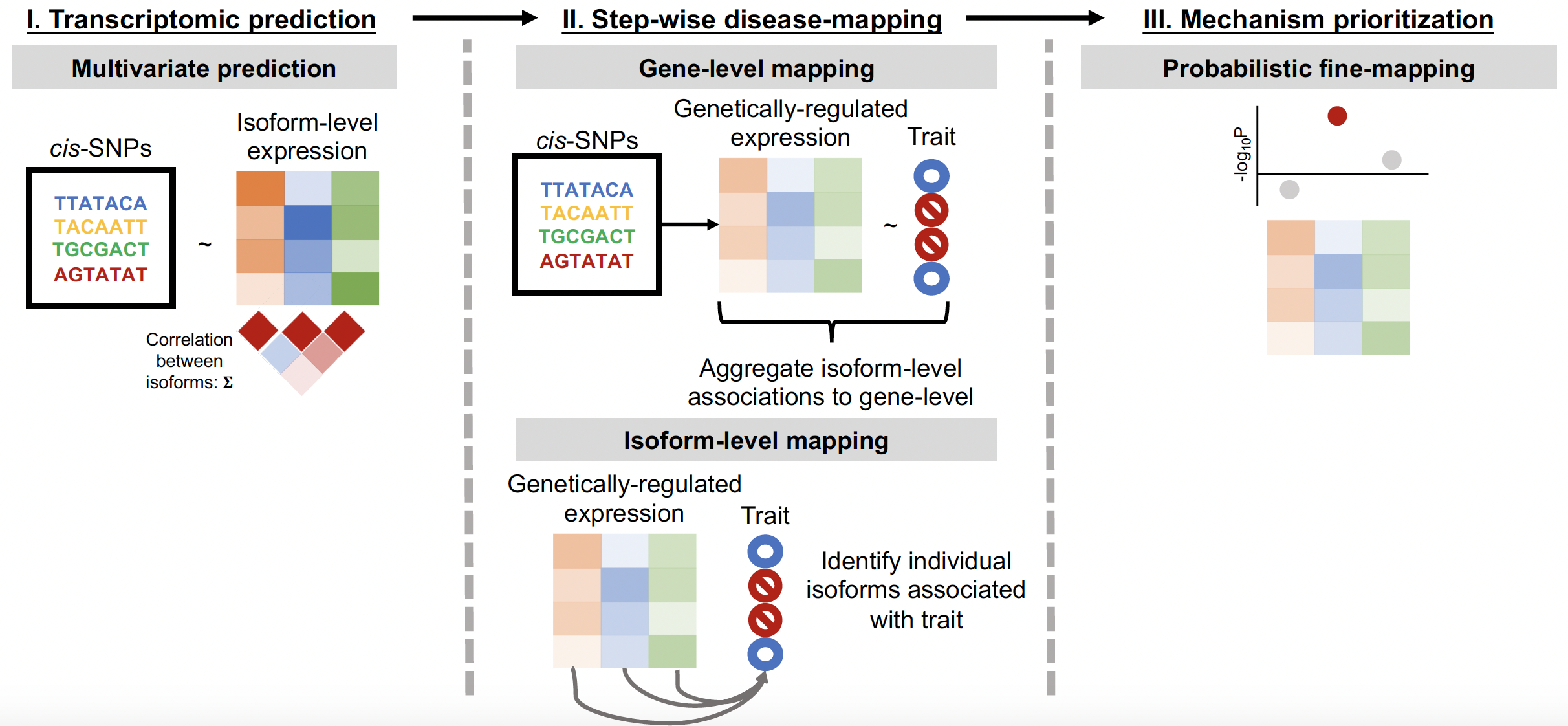
Conducting trait mapping on the isoform-level using stage-wise hypothesis testing
Probabilistic fine-mapping to identify causal isoforms
Key Features
Multiple prediction methods: The package automatically evaluates multiple multivariate regression methods and selects the best model based on cross-validated R-squared.
Graph-regularized learning: Incorporate prior knowledge about isoform similarity (e.g., shared exon structure) to improve predictions.
User-friendly interface: The main
compute_isotwas()function provides detailed progress output, data summaries, and results comparison tables.Robust model selection: Uses cross-validation within each method and compares across methods to find the optimal model for each gene.
See the Train an isoTWAS model vignette for detailed usage instructions.